Annotation as a service: helping build a global knowledge layer with annotation of the dark literature.¶
US Team Lead: C. Titus Brown, UC Davis.
non-US Team Lead: Christof Dorner, Hypothes.is.
Introduction:¶
Hypothes.is is an annotation overlay that allows annotations to be associated with Web text. Each annotation consists of a document ID, a document anchor, some free text, and tags; annotations can be either public or belong to private groups. The Hypothes.is service currently offers an overlay display where these annotations are placed on HTML or PDF views of documents; annotations can also be shared and searched globally. Annotations can be placed manually by means of a browser plugin, or the same code delivered via JavaScript embedding or a Web proxy. Annotations are stored in the Hypothes.is central database and can be retrieved on demand for any specified URL, user, group, or tag. Annotations are searchable along these facets as well as with free text query. Annotations will soon be link-able to ORCIDs, as well, tying into a global system of authorship identifiers.
Hypothes.is offers an optional overlay and enables permissionless annotation by 3rd parties unaffiliated with either the content consumer or the content provider. Crucially, this bypasses much of the friction associated with working with publishers to provide an annotation overlay, although a consortium of publishers is working closely with Hypothes.is.
For more on Hypothes.is, please see Introducing Hypothes.is.
The problems and the proposed solution¶
While deceptively simple in concept, the potential of Hypothes.is to provide a dynamic unifying layer across biomedicine is significant.
There are two problems we are interested in solving.
First, large portions of the biomedical scholarly literature are completely inaccessible to automated, large scale analysis. Moreover, large portions of the biomedical literature may have been annotated, either by humans or automated algorithms, but these annotations are not easily shared, nor are they available for global analysis. Rather, each is locked in an individual silo.
Second, the Hypothes.is annotation system currently relies on manual annotation.
We believe we can help solve these two bigger problems with one technical approach: we will give individuals an in-browser app letting them submit any paper to an analysis server that will openly annotate the paper. We will also build server-side software to support any annotation engine.
This will permit individuals to link closed papers of interest into the global annotation web, while also enabling new annotation services to be built quickly and easily.
More generally, this can drive the creation of machine-based structured tags to enrich texts, which will create a skein of linked data that could be surfaced in various ways. And, because annotations are RDF, the recording and exposure of data linkages becomes available for querying. In essence, it would help create derived knowledge that could be queried in powerful ways.
We have provisionally named this server-side system “Aesir.”
A prototype: SciBot¶
Working with partners at the Neuroscience Information Framework (NIF), Hypothes.is has deployed an alpha service called SciBot. SciBot is a tool that scans scientific papers for text snippets called RRIDs (Research Resource Identifiers), annotates them with metadata stored elsewhere, and enables human curators to validate or enhance the connection between an RRID that appears in the text of a paper and its associated metadata. RRID are unique identifiers for certain types of resources used in biomedicine, e.g. antibodies, model organisms and software tools/databases. These identifiers are issued by authoritative community repositories like the Antibody Registry, and aggregated by NIF through its data aggregation platform, SciCrunch. When the SciBot plug in is activated, it highlights RRID’s in the text and pulls the metadata about the particular resource from the SciCrunch resolving service into the Hypothes.is client, where it is presented as an annotation overlay on the text. Viewers can reply to the annotation, e.g., to note an error or supply additional information on a resource. In this way, Hypothes.is creates a hybrid machine-human interface that has general purpose utility. Aesir would extend this to arbitrary annotation engines.
Please see the associated video for a demo. The source code is publicly available under an open source license at https://github.com/judell/rrid.
Three use cases that Aesir would enable¶
Annotating closed biomedical literature with links to genome databases. Rebecca is reading a C. elegans paper in a closed-access journal, and would like to decorate it with annotations connecting genes and genome coordinates to Wormbase, the C. elegans genome database. She installs the Wormbase “annotate this” bookmark and clicks on it; this sends the paper to an Aesir server that annotates it and posts the annotations to Hypothes.is, where they can be viewed by Rebecca and others.
This is a general pattern: many databases may want to layer annotations on the literature, but working with many publishers directly can be an insurmountable challenge. Hypothes.is solves this problem, but as of yet there is no good way for databases to engage with Hypothes.is in high-throughput.
Extract data from closed literature. Jason would like to use ContentMine to extract data from a closed-access paper he is reading, for his own reuse. He clicks the ContentMine “extract this” bookmark, which sends the paper to an Aesir server which runs the ContentMine annotation engine on it and redirects Jason to the resulting data. In line with ContentMine’s collaboration with Hypothes.is, links to the data are also provided directly on the paper.
By building a system that lets users submit interesting literature to the annotation system, we believe we can crack open the closed literature to an significant extent. We also expect a higher density of annotations around more “active” or still-relevant papers, which will provide interesting opportunities for meta-analysis.
Leveraging & driving open peer review. Tina is reviewing a paper, and wants to mark it up with external blog commentary and public reviews. Unfortunately, the paper is a revision of a posted preprint and so its Hypothes.is URL has changed and no annotations are associated with it; James submits the paper to an Aesir-based service that identifies it as a duplicate of a previously annotated paper and transfers the annotations over to it where possible. As a bonus, James could annotate the paper with his review when he is done.
Here, Hypothes.is could help tame the impending chaos of pre- and post-publication open peer review, by interlinking comments from multiple sources to each paper. These comments could even be summarized and overlaid by a meta-analysis service, providing additional value to readers.
In these three use cases, considerable computational work is being done by the remote server - none of this work can be done within the confines of the current client-side Hypothes.is approach, and all three use cases could be supported with a single technical solution.
Deliverables¶
The first line of deliverables would be a full server (implemented in Python) for adding annotations to documents, together with JavaScript bookmarklets to enable submission of documents to such servers from within any modern Web browser. The server code would contain an open source implementation of the parsing and annotation-marking logic used in Hypothes.is, providing a framework that would readily enable developers to consume text and annotate it according to their own logic. We would provide a full demo server implementation using a public open source code base (here, working with ContentMine would be a natural fit). We would also provide simple hooks to enable anyone to integrate whatever annotation engine they wanted.
On top of these deliverables we envision a variety of services, depending on where we see opportunities emerge:
- large-scale annotation search and comparison, so that documents can be grouped and analyzed based on their annotations and larger networks of annotations;
- enabling human feedback on and editing of automated annotations within Hypothes.is itself, so that viewers can annotate annotations and identify misannotations;
- a recommendation system based on annotation similarity and interlinking, so that when annotations become dense enough, new associates can be found between existing literature;
- a notification service that would allow people to “watch” collections of annotations and/or publications, and filter the notifications;
- more advanced annotation overlays for documents;
- tools for managing annotations from multiple sources, integrating with social information (lab groups, collaborators, institutions), and support for trust network overlays;
- unifying scattered versions and representations by marrying automatic analysis and human curation; for example, Hypothes.is could be used to store “document signatures” that could be searched to connect papers with different URIs.
Impacts and ecosystem¶
First and foremost, we believe we can enable an ecosystem of annotation algorithms, some general (e.g. linguistic analysis of document structure) and some field-specific (e.g. automated extraction and annotation of drug-drug interactions across document collections).
Second, we envision this as providing entree to the “dark literature” via annotations. Annotations are exempt from publisher’s usage restrictions, which means that anyone with access to a document can submit it to an annotation server, layer annotations upon the document, and publicize these annotations.
Third, by integrating human feedback and meta-annotation of these primary annotations, we can help provide algorithm authors with “eyeballs” and corrections on their annotations. This kind of feedback will help drive better annotation algorithms.
Fourth, with tools to compare collections of annotations across large bodies of papers, we will enable meta-analysis of annotation networks and be able to build connections between overlapping but disparate subfields of scientific literature.
Fifth, we can provide tools for databases, publishers and libraries to work with closely held document collections. For example, someone with access to large bodies of papers could annotate with the results of data mining applied to the paper; or, users with institutional access to closed-access publications could submit those publications to a ContentMine server for open annotation; or, biological databases could use their special knowledge of identifiers to provide annotations based on a compute-intensive analysis; or, publishers and institutions could provide a “first look” of annotation for their own document collections; or, reviewers could privately annotate papers they are reviewing with an automated system to identify relevant literature.
Why does this fit the Open Science Prize?¶
We are encouraging and supporting services, tools, and platforms that enable the generation of open content: here, the comments, extractions, and annotations on literature. The proposed system could also consume, integrate, analyze, and compare existing annotations, enabling meta-analysis of annotations. And, since annotations in Hypothes.is are under CC0, there is no restriction on reuse or remixing.
Moreover, the use of closed access literature to produce annotations is arguably legal because it is non-infringing transformative fair use (although this has not been tested yet; see link for argument). By making use of remote “cloud” services to analyze (but not retain) literature and doing so at a user’s request, we believe we avoiding any infringement of license terms. We do expect that the initial beneficiaries of this will be the closed-access publishers who may find more users of their closed archives, but even this serves the greater good by linking this closed literature into the global annotatome. In the long term, we believe this will serve as a powerful demonstration of the uses and power of open literature.
Longer term, we believe there will be many groups interested in permissionless server-side automated annotation of text, and we hope that by providing an automated system matching the existing client annotation interface, we can more readily enable the development of an ecosystem of approaches to open annotation. Essentially, we hope to expand the range of creative people who can try out new annotation approaches.
Additional use cases¶
Duplication, version, and plagiarism analysis could be performed automatically. It would be straightforward to identify cases where highly similar annotations were placed on different document IDs; the documents could then be examined for document equivalence, different versions, or plagiarized text.
Annotations could enable back citation from future literature, including identification of retracted citations, comments, and blog posts on the work and derived works.
Hypothes.is could be used to place forward links to software, computational workflows, & replications of published work on papers automatically.
What would we spend the money on?¶
The prize money would be spent on developing an open source server and proof of concept, and applying it to our three primary use cases. All software products would be under the BSD 3-clause license. Hypothes.is itself is completely open source.
We plan to support an existing Hypothes.is developer to implement the prototype, and will then run hackathons once we have an initial proof of concept.
Architecture¶
Our primary client-side mechanism will be a bookmarklet or in-browser app that would submit HTML or PDF text to a server for analysis. On the server side, we would provide Python libraries for text consumption and normalization, anchor extraction, annotation retrieval, and annotation submission. Our ultimate goal is to open up an ecosystem of annotation production, consumption, and meta-analysis to everyone, and to allow anyone with access to any document to enter it into this ecosystem.
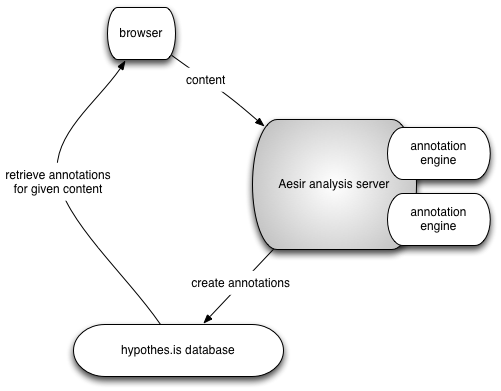
Figure 1: The basic architecture we propose; the annotation engines would make use of but be largely independent of the Aesir code.
LICENSE: This page and all textual/graphic site content is licensed under the Creative Commons - 0 License (CC0).
comments powered by Disqus